behavioural
epigenetics

The author, Margaret Tyson BSc MPHe PhD , was an honorary researcher at the Institute of Cancer Sciences, The University of Manchester and now researches epigenetics particularly of cancer and schizophrenia. She also runs Manchester Amputee Fitnesss Initiative
 |
The main aim of this website
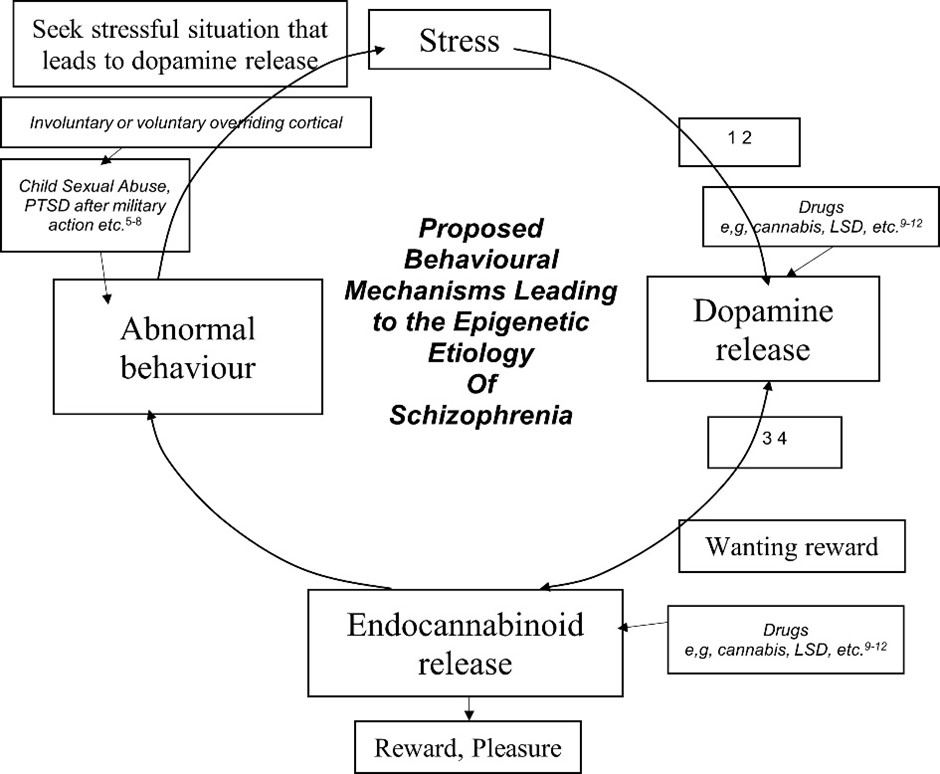
Previously, it has been thought that neurons in the brain were unaltered after infantile pruning and that we gradually lost them as we aged. Genes were seen as being the only way that behaviour was formed. Since the study of the epigenetic modification of genes has been a growing discipline it has been realised that epigenetic change is important in the study of serious diseases especially in mental health. Genes can be changed by our environment but also by our behaviour. This can result in diseases such as schizophrenia/psychosis. There is now evidence that it is behaviour which causes schizophrenia through epigenetic change to the brain.
While abnormal behaviour may cause schizophrenia, positive behaviour such as learning a new skill or training and being in a rich environment results in positive effects on the brain such as increased grey matter volume (intellect) again through epigenetic change to the brain. These pages are based on a review I am writing and I will keep updating these pages as I research the subject.
This page details the epigenetic mechanisms that change genes. However, for information about these effects on the body specifically the brain due to abnormal behaviour which may result in illnesses particularly schizophrenia read the following pages.
Epigenetic mechanisms
Just a short description so that people will understand that it’s not just about genes!
Up until recently it was thought that mammalian blue print, in this case human, consisted of the genome (the total collection of our genes) i.e. characteristics inherited from our parents which is fixed. Increasingly it is understood that these genes can be altered by the environment, that is, what we are exposed to e.g. our diet, exercise, chemicals and even our behaviour.
The ENCODE human genome project revealed that “junk DNA” is no longer a fact and that although only about 20% of DNA encodes for proteins the rest encodes for many types of regulatory RNAs, histone modification, gene modification such as methylation etc. (1, 2)
There are several different ways that a genome can be changed or modified by epigenetic mechanisms (3). There is complex cross-talk between these modifications so that everything we encounter is transcribed and translated into our bodies. The main mechanisms are: methylation, acetylation and microRNA (miRNA) control (4-6).
1. Methylation 2
DNA Methylation (Fig 1) is mainly an inhibitory mechanism where a methyl group is added reducing a gene’s function. The opposite is also used to reinstate a gene’s function (demethylation).
2. Acetylation 2
Histone Acetylation (Fig 1) usually enhances gene activity through various mechanisms including triggering the removal of a methyl group.
3. MiRNA control (2+5)
MicroRNAs (MiRNA) are small non-coding RNA molecules which attach themselves to sections of mRNA (messenger RNA) to block translation of the underlying DNA to proteins (Fig 2). There is an feedback relationship between MiRNA control and the epigenetic modifications shown above (7). This is essential to the normal functioning of cells. When this is faulty it can lead to serious diseases such as cancer, cardiovascular disease and schizophrenia.
A good website that explains epigenetics is:
Fig 1 From: Vilcinskas A. The role of epigenetics in host–parasite coevolution: lessons from the model host insects Galleria mellonella and Tribolium castaneum. Zoology 2016;119(4):273-80. doi: https://doi.org/10.1016/j.zool.2016.05.004
Fig. 2 MiRNA blockage of messenger RNA (mRNA) translation to proteins (from naturamatematica.blogspot.com)
References
1. Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science 2012;337(6099):1159, 61. doi: 10.1126/science.337.6099.1159 [published Online First: 2012/09/08]
2. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489(7414):57-74. doi: 10.1038/nature11247
3. Handy DE, Castro R, Loscalzo J. Epigenetic Modifications: Basic Mechanisms and Role in Cardiovascular Disease. Circulation 2011;123(19):2145-56. doi: 10.1161/CIRCULATIONAHA.110.956839
4. Moore LD, Le T, Fan G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013;38(1):23-38. doi: 10.1038/npp.2012.112
5. Chaturvedi P, Tyagi SC. Epigenetic mechanisms underlying cardiac degeneration and regeneration. International Journal of Cardiology 2014;173(1):1-11. doi: https://doi.org/10.1016/j.ijcard.2014.02.008
6. Vilcinskas A. The role of epigenetics in host–parasite coevolution: lessons from the model host insects Galleria mellonella and Tribolium castaneum. Zoology 2016;119(4):273-80. doi: https://doi.org/10.1016/j.zool.2016.05.004
7. Liep J, Rabien A, Jung K. Feedback networks between microRNAs and epigenetic modifications in urological tumors. Epigenetics 2012;7(4):315-25. doi: 10.4161/epi.19464